5.13 MCMC for DINA model in Nimble R package
Code
library(nimble)
library(GDINA)
library(MCMCvis)
library(nimble, warn.conflicts = FALSE)
DINA.mcmc <- nimbleCode({
for (n in 1:N) {
for (j in 1:J) {
prob[n, j] <- g[j] + (1 - s[j] - g[j]) * 1 * (sum(alpha[n,
1:K] * Q[j, 1:K]) >= sum(Q[j, 1:K]))
score[n, j] ~ dbern(prob[n, j])
}
latent.group.index[n] ~ dcat(pi[1:C])
alpha[n, 1:K] <- all.patterns[latent.group.index[n], 1:K]
}
pi[1:C] ~ ddirch(delta[1:C])
for (j in 1:J) {
s[j] ~ dbeta(1, 1)
g[j] ~ T(dbeta(1, 1), , 1 - s[j])
}
})
score <- sim10GDINA$simdat
Q <- sim10GDINA$simQ
J <- ncol(score)
K <- ncol(Q)
N <- nrow(score)
all.patterns <- GDINA::attributepattern(K)
C <- nrow(all.patterns)
DINAconstants <- list(Q = Q, N = N, J = J, K = K, C = C, delta = rep(1,
C))
DINAdata <- list(score = score, all.patterns = as.matrix(all.patterns))
initials <- list(list(s = rep(0.2, J), g = rep(0.2, J), pi = rep(1/C, C),
latent.group.index = sample(1:C, N, replace = TRUE)), list(s = rep(0.1,
J), g = rep(0.1, J), pi = rep(1/C, C), latent.group.index = sample(1:C,
N, replace = TRUE)))
nimbleMCMC_samples <- nimbleMCMC(code = DINA.mcmc, constants = DINAconstants,
data = DINAdata, inits = initials, monitors = c("s", "g"), nburnin = 2500,
niter = 5000, nchains = 2, setSeed = c(123, 456))Code
library(MCMCvis)
MCMCvis::MCMCsummary(nimbleMCMC_samples, params = c("s", "g"), Rhat = TRUE,
round = 2)## mean sd 2.5% 50% 97.5% Rhat n.eff
## s[1] 0.26 0.03 0.20 0.26 0.31 1.0 306
## s[2] 0.28 0.03 0.23 0.28 0.33 1.0 408
## s[3] 0.11 0.02 0.07 0.11 0.16 1.0 308
## s[4] 0.20 0.03 0.14 0.20 0.25 1.0 334
## s[5] 0.29 0.04 0.22 0.29 0.36 1.0 370
## s[6] 0.07 0.02 0.04 0.07 0.11 1.0 493
## s[7] 0.32 0.03 0.26 0.32 0.37 1.0 597
## s[8] 0.31 0.04 0.23 0.31 0.37 1.0 248
## s[9] 0.21 0.03 0.16 0.21 0.26 1.0 665
## s[10] 0.16 0.03 0.10 0.16 0.22 1.0 585
## g[1] 0.07 0.05 0.00 0.07 0.17 1.0 66
## g[2] 0.09 0.03 0.04 0.09 0.15 1.0 220
## g[3] 0.07 0.03 0.03 0.07 0.13 1.0 192
## g[4] 0.22 0.02 0.18 0.22 0.26 1.0 517
## g[5] 0.08 0.01 0.06 0.08 0.11 1.0 809
## g[6] 0.59 0.03 0.53 0.59 0.63 1.0 533
## g[7] 0.26 0.02 0.21 0.26 0.30 1.0 683
## g[8] 0.15 0.02 0.11 0.15 0.19 1.0 449
## g[9] 0.27 0.02 0.23 0.27 0.31 1.0 723
## g[10] 0.33 0.02 0.30 0.33 0.37 1.0 690Code
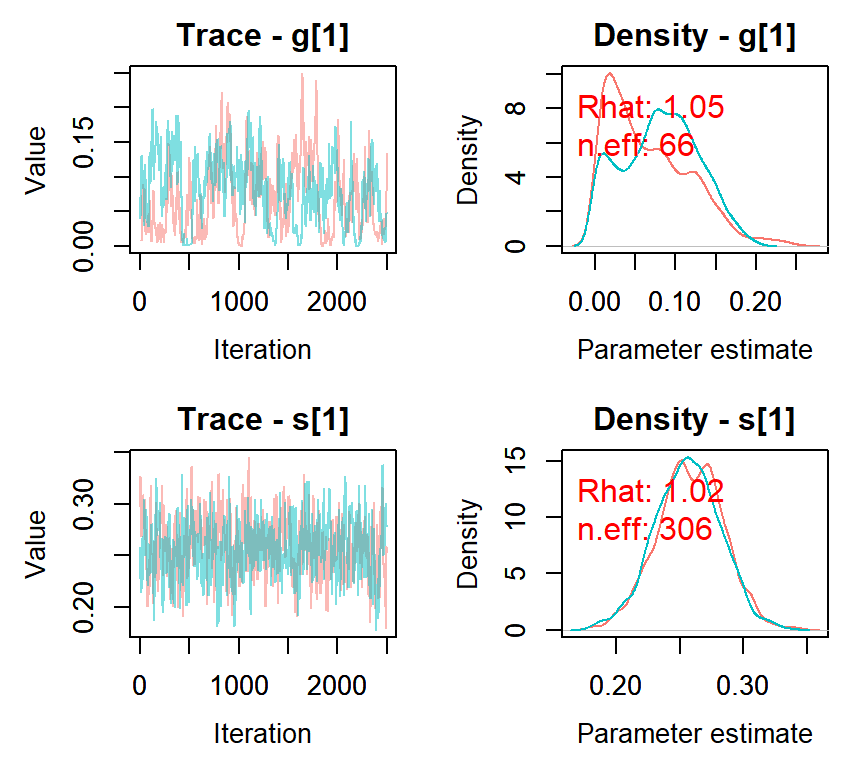
MCMCvis::MCMCtrace(nimbleMCMC_samples, params = c("g[1]", "s[1]"), ISB = FALSE,
exact = TRUE, ind = TRUE, pdf = FALSE, Rhat = TRUE, n.eff = TRUE)